iUSEiSEE - Bonus content
Federico
Marini1,
Najla Abassi2
Collection of material developed by the iSEE core dev team
(Kevin
Rue-Albrecht, Federico Marini, Charlotte Soneson, Aaron Lun)
Source: vignettes/bonus_content_04.Rmd
bonus_content_04.Rmd
Moar iSEE things
This vignette contains some examples on even more
iSEE-things (Rue-Albrecht et al.
2018).
We will use the following packages throughout its content (make sure to have them installed if you want to run it interactively)
iSEE a challenge: let’s reproduce any figure!
We’d like to open up a challenge, with some simple rules:
- You search a nice graphical representation of data
- We try to reproduce this within
iSEE- as far as this is possible!
Let’s suggest a couple of figure/figure panels!
Enter a couple of suggestions in this GSheet: https://docs.google.com/spreadsheets/d/1poE713rXqzfNdPcKAPN2AxJYXhaxy8GwgAjRsk_UiK8/edit?usp=sharing
iSEEde and iSEEpathways
iSEEde and iSEEpathways: ideal companions for exploring DE results
We first load the processed macrophage data (derived
from the work of (Alasoo et al. 2018)) -
on this, we already ran the workflow of DESeq2 (Love, Huber, and Anders 2014) to identify the
differentially expressed genes.
macrophage_location <- system.file("datasets", "sce_macrophage_readytouse.RDS",
package = "iUSEiSEE"
)
macrophage_location
#> [1] "/Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/library/iUSEiSEE/datasets/sce_macrophage_readytouse.RDS"
sce_macrophage <- readRDS(macrophage_location)
library("iSEE")
library("iSEEde")
library("iSEEpathways")iSEEde and
iSEEpathways
are two new Bioconductor packages that provide iSEE panels
specifically aimed towards exploration of differential expression and
pathway analysis results. More precisely, iSEEde
provides the VolcanoPlot, MAPlot,
LogFCLogFCPlot and DETable panels. These
panels can be configured to extract data that was added via the
embedContrastResults() function above. Let’s look at an
example:
app <- iSEE(sce_macrophage, initial = list(
DETable(
ContrastName = "IFNgTRUE.SL1344TRUE.DESeq2",
HiddenColumns = c("baseMean", "lfcSE", "stat")
),
VolcanoPlot(ContrastName = "IFNgTRUE.SL1344TRUE.DESeq2"),
MAPlot(ContrastName = "IFNgTRUE.SL1344TRUE.DESeq2")
))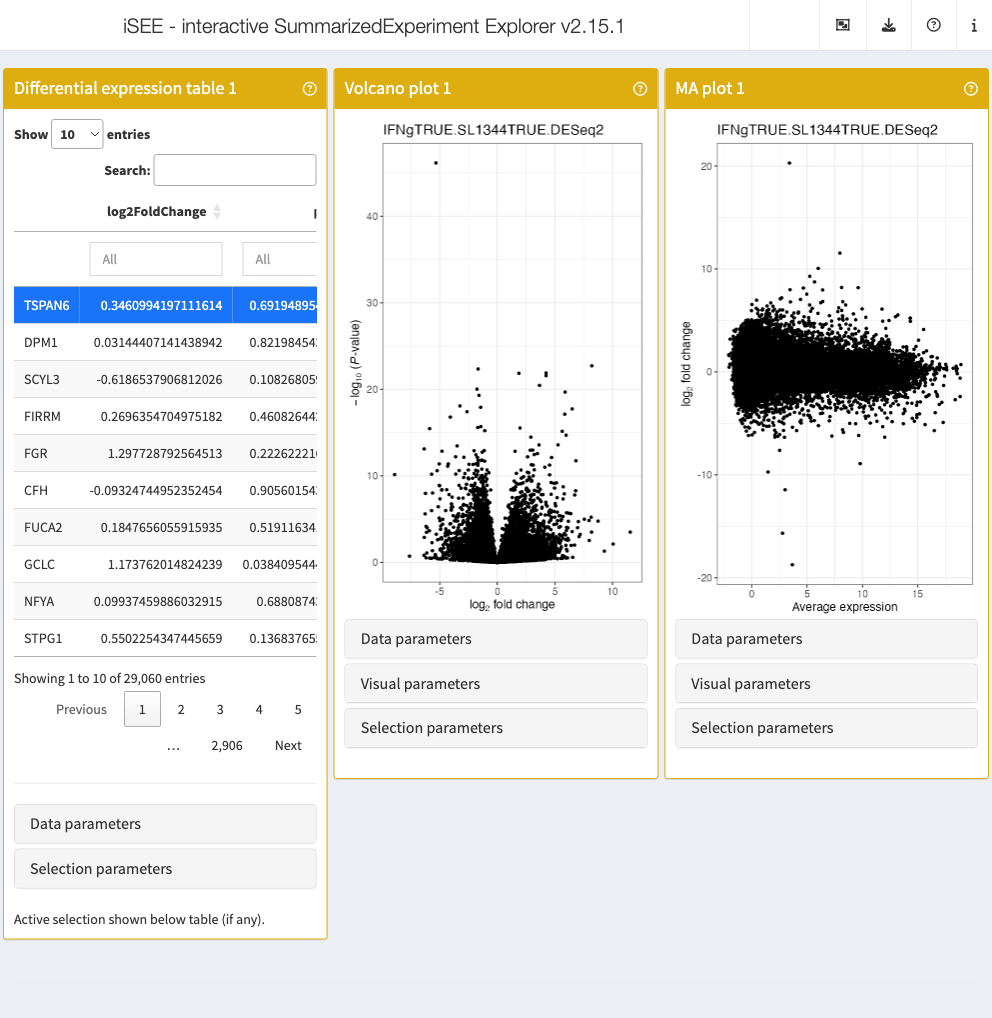
Note how it is easy to switch to a different contrast in any of the panels.
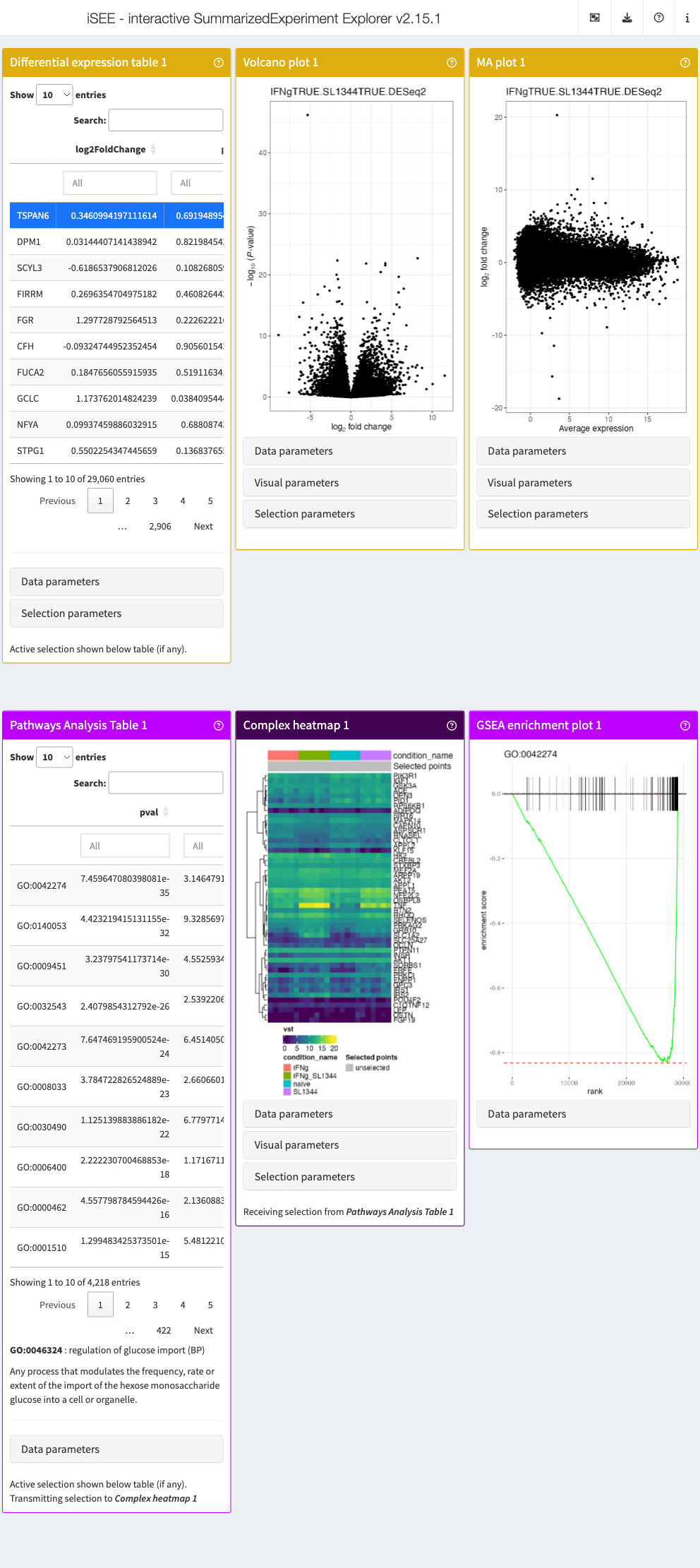
app <- iSEE(sce_macrophage, initial = list(
iSEEde::DETable(
ContrastName = "IFNgTRUE.SL1344TRUE.DESeq2",
HiddenColumns = c("baseMean", "lfcSE", "stat")
),
iSEEde::VolcanoPlot(ContrastName = "IFNgTRUE.SL1344TRUE.DESeq2"),
iSEEde::MAPlot(ContrastName = "IFNgTRUE.SL1344TRUE.DESeq2"),
PathwaysTable(
ResultName = "IFNgTRUE.SL1344TRUE.limma.fgsea",
Selected = "GO:0046324"
),
ComplexHeatmapPlot(
RowSelectionSource = "PathwaysTable1",
CustomRows = FALSE, ColumnData = "condition_name",
ClusterRows = TRUE, Assay = "vst"
),
FgseaEnrichmentPlot(
ResultName = "IFNgTRUE.SL1344TRUE.limma.fgsea",
PathwayId = "GO:0046324"
)
))
iSEEfier
Let’s say we are interested in visualizing the expression of a list of specific marker genes in one view, or maybe we created different initial states separately, but would like to visualize them in the same instance. As we previously learned, we can do a lot of these tasks by running the command:
# don't run
iSEE(sce)Then, add/remove the panels to our need. This can involve multiple
steps (selecting the gene of interest, color by a specific
colData…), or probably even write multiple lines of code.
For that we can use the iSEEfier
package, which streamlines the process of starting (or if you will,
firing up) an iSEE instance with a small chunk of code,
avoiding the tedious way of setting up every iSEE panel
individually.
In this section, we will illustrate a simple example of how to use iSEEfier. We will use the same pbmc3k data we worked with during this workshop.
We start by loading the data:
library("iSEEfier")
# import data
sce <- readRDS(
file = system.file("datasets", "sce_pbmc3k.RDS", package = "iUSEiSEE")
)
sce
#> class: SingleCellExperiment
#> dim: 32738 2643
#> metadata(0):
#> assays(2): counts logcounts
#> rownames(32738): MIR1302-10 FAM138A ... AC002321.2 AC002321.1
#> rowData names(19): ENSEMBL_ID Symbol_TENx ... FDR_cluster11
#> FDR_cluster12
#> colnames(2643): Cell1 Cell2 ... Cell2699 Cell2700
#> colData names(24): Sample Barcode ... labels_ont cell_ontology_labels
#> reducedDimNames(3): PCA TSNE UMAP
#> mainExpName: NULL
#> altExpNames(0):For example, we can be interested in visualizing the expression of GZMB, TGFB, and CD28 genes all at once. We start by providing a couple of parameters:
# define the list of genes
feature_list_1 <- c("GZMB", "TGFB1", "CD28")
# define the cluster/cell type
cluster_1 <- "labels_main"Now we can pass these parameters into iSEEinit() to
create a customized initial configuration:
# create an initial state with iSEEinit
initial_1 <- iSEEinit(sce,
features = feature_list_1,
clusters = cluster_1,
add_markdown_panel = TRUE)The rest can be as easy as passing this initial to the
iSEE() call:
app <- iSEE(sce, initial = initial_1)This is how it would look like:
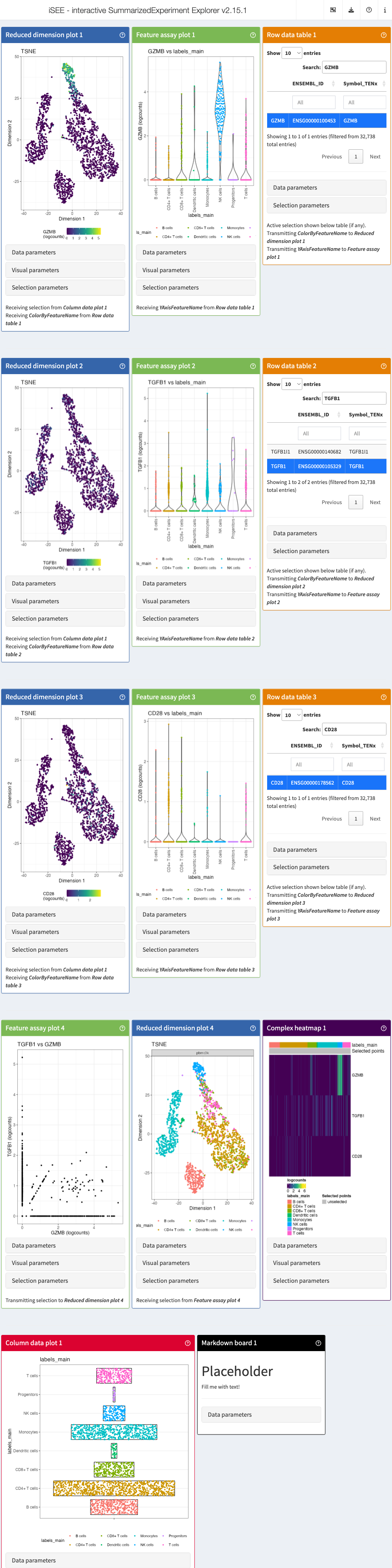
While we are visualizing the expression of these genes, we might want
to take some notes (gene X is more expressed in a certain cell
type/cluster than some others, maybe we are trying to annotate the cells
ourselves if the annotation wasn’t available…).
For this, we used the argument add_markdown_panel = TRUE.
It will display a MarkdownBoard panel where we can note our
observations without leaving the app.
We can also use the argument add_dynamicTable_panel=TRUE
to add another custom panel to display the maker genes of certain
cluster/cell type.
feature_list_2 <- c("CD74", "CD79B")
initial_2 <- iSEEinit(sce,
features = feature_list_2,
clusters = cluster_1,
add_markdown_panel = TRUE,
add_dynamicTable_panel = TRUE)
app <- iSEE(sce, initial = initial_2)We can check the initial’s content, or how the included panels are
linked between each other without running the app with
view_initial_tiles() and
view_initial_network():
# display a graphical representation of the initial configuration, where the panels are identified by their corresponding colors
view_initial_tiles(initial = initial_1)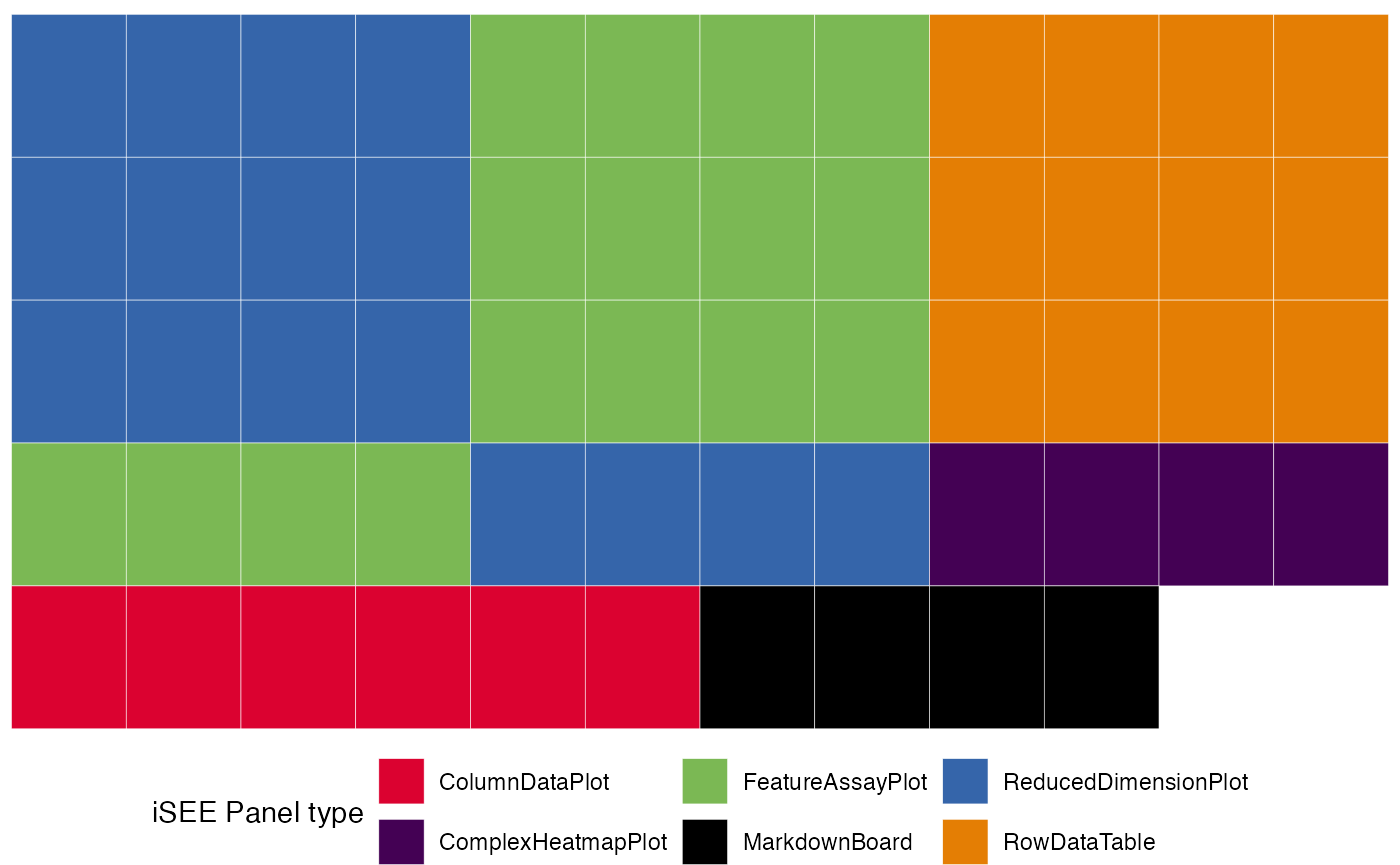
# display a network visualization for the panels
view_initial_network(initial_1, plot_format = "igraph")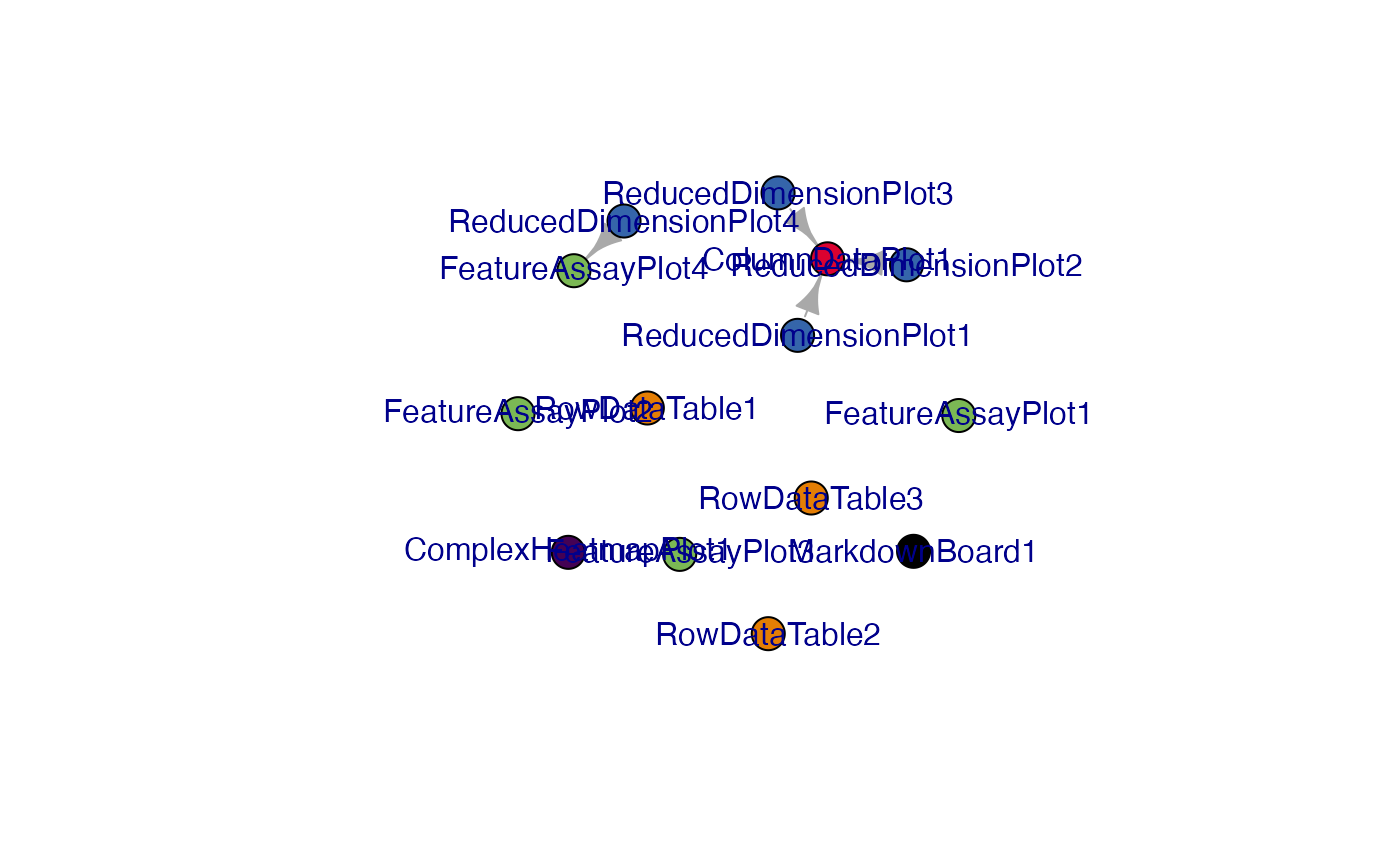
#> IGRAPH edf2d5a DN-- 14 4 --
#> + attr: name (v/c), color (v/c)
#> + edges from edf2d5a (vertex names):
#> [1] ReducedDimensionPlot1->ColumnDataPlot1
#> [2] ReducedDimensionPlot2->ColumnDataPlot1
#> [3] ReducedDimensionPlot3->ColumnDataPlot1
#> [4] ReducedDimensionPlot4->FeatureAssayPlot4Another alternative for network visualization would use the
interactive widget provided by visNetwork:
view_initial_network(initial_1, plot_format = "visNetwork")
#> IGRAPH f1b0726 DN-- 14 4 --
#> + attr: name (v/c), color (v/c)
#> + edges from f1b0726 (vertex names):
#> [1] ReducedDimensionPlot1->ColumnDataPlot1
#> [2] ReducedDimensionPlot2->ColumnDataPlot1
#> [3] ReducedDimensionPlot3->ColumnDataPlot1
#> [4] ReducedDimensionPlot4->FeatureAssayPlot4It is also possible to combine multiple initials into one:
merged_config <- glue_initials(initial_1, initial_2)
#> Merging together 2 `initial` configuration objects...
#> Combining sets of 14, 14 different panels.
#>
#> Dropping 2 of the original list of 28 (detected as duplicated entries)
#>
#> Some names of the panels were specified by the same name, but this situation can be handled at runtime by iSEE
#> (This is just a non-critical message)
#>
#> Returning an `initial` configuration including 26 different panels. Enjoy!
#> If you want to obtain a preview of the panels configuration, you can call `view_initial_tiles()` on the output of this function
# check out the content of merged_config
view_initial_tiles(initial = merged_config)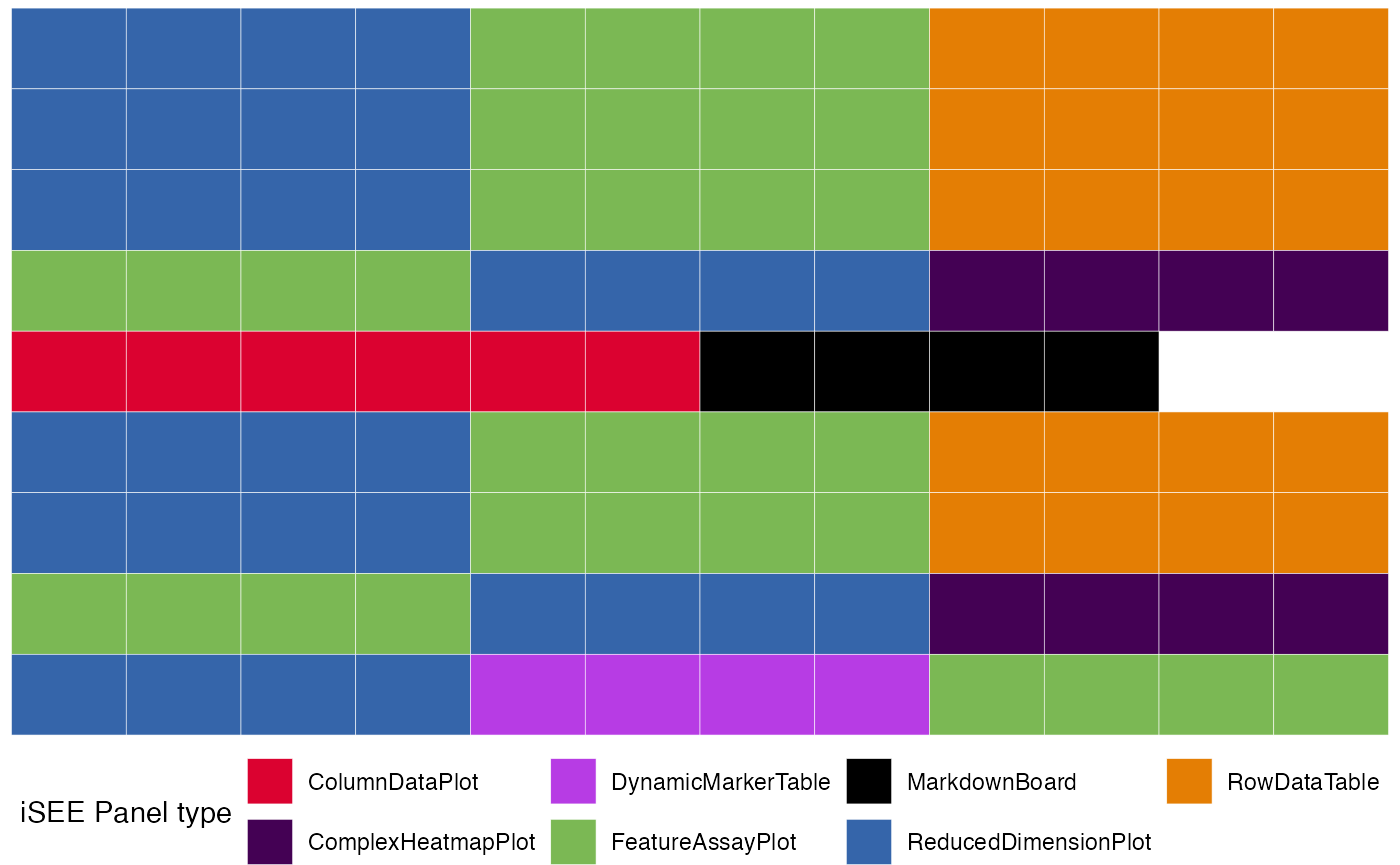
?iSEEfier is always your friend whenever you need
further documentation on the package/a certain function and how to use
it.
Multi-sample, multi-condition datasets: how to let iSEE shine
We will take another dataset for this, from a recent work of Bartneck and colleagues ((Bartneck et al. 2023)).
You can find the dataset in the sce_merged_allsets.RDS
object - let’s see together how we can access that –> https://seafile.rlp.net/d/c7f1335a6248427d97da/.
Once the data is available locally, we load it in the R session and simply start iSEE on it.
For more about this work, please refer to the original publication.
iSEEhub & iSEEindex
iSEEhub: iSEEing the ExperimentHub datasets
The iSEEhub package provides a custom landing page for an iSEE application interfacing with the Bioconductor ExperimentHub. The landing page allows users to browse the ExperimentHub, select a data set, download and cache it, and import it directly into an iSEE app.
library("iSEE")
library("iSEEhub")
#> Loading required package: ExperimentHub
#> Loading required package: AnnotationHub
#> Loading required package: BiocFileCache
#> Loading required package: dbplyr
#>
#> Attaching package: 'AnnotationHub'
#> The following object is masked from 'package:Biobase':
#>
#> cache
ehub <- ExperimentHub()
app <- iSEEhub(ehub)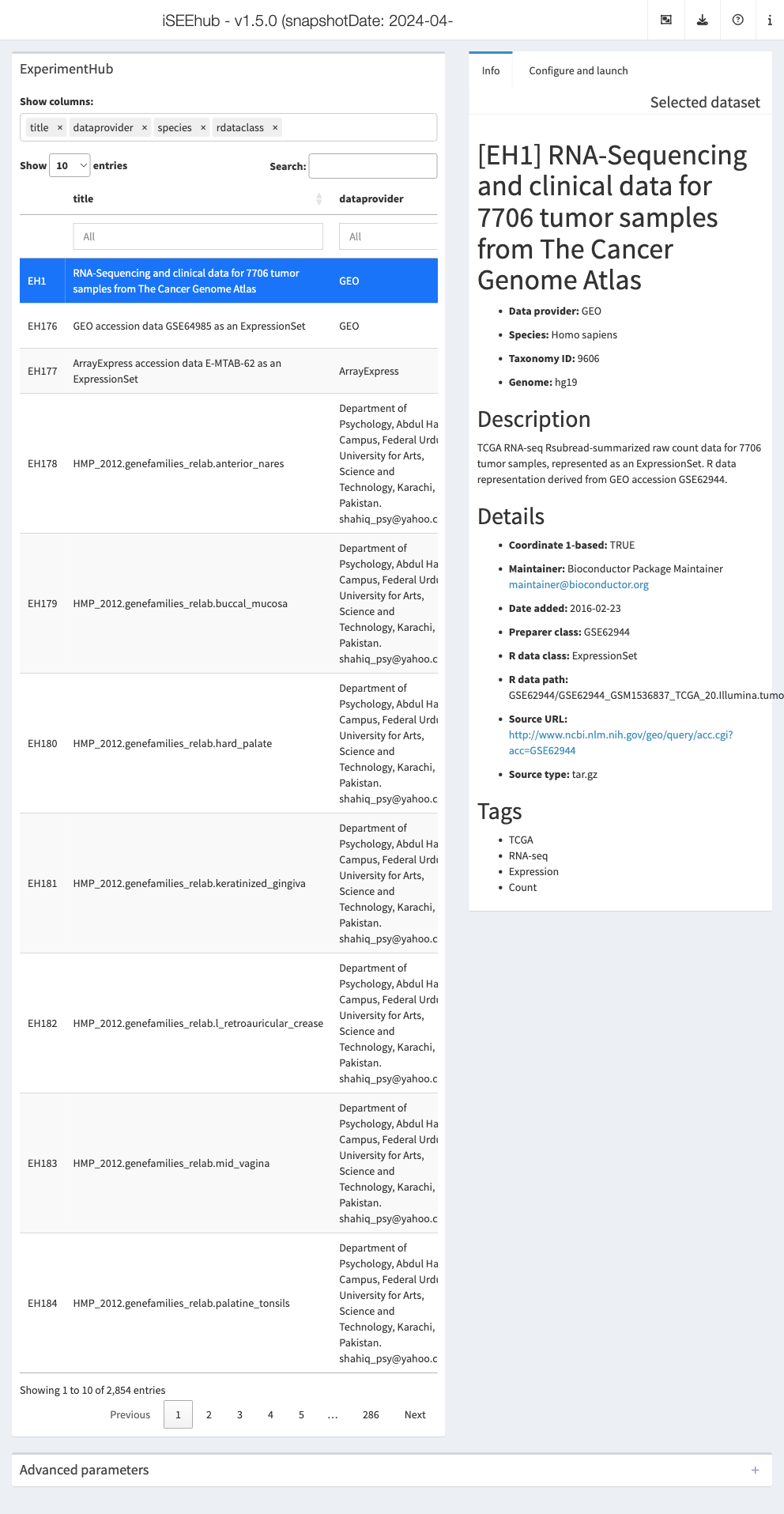
iSEEindex: one instance of iSEE to explore them
all
iSEEindex
provides an interface to any collection of data sets
within a single iSEE web-application.
The main functionality of this package is to define a custom landing
page allowing app maintainers to list a custom collection of data sets
that users can select from and directly load objects into an iSEE web
application. To see how to configure such an app, we will create a small
example:
library("iSEE")
library("iSEEindex")
bfc <- BiocFileCache(cache = tempdir())
dataset_fun <- function() {
x <- yaml::read_yaml(system.file(package = "iSEEindex", "example.yaml"))
x$datasets
}
initial_fun <- function() {
x <- yaml::read_yaml(system.file(package = "iSEEindex", "example.yaml"))
x$initial
}
app <- iSEEindex(bfc, dataset_fun, initial_fun)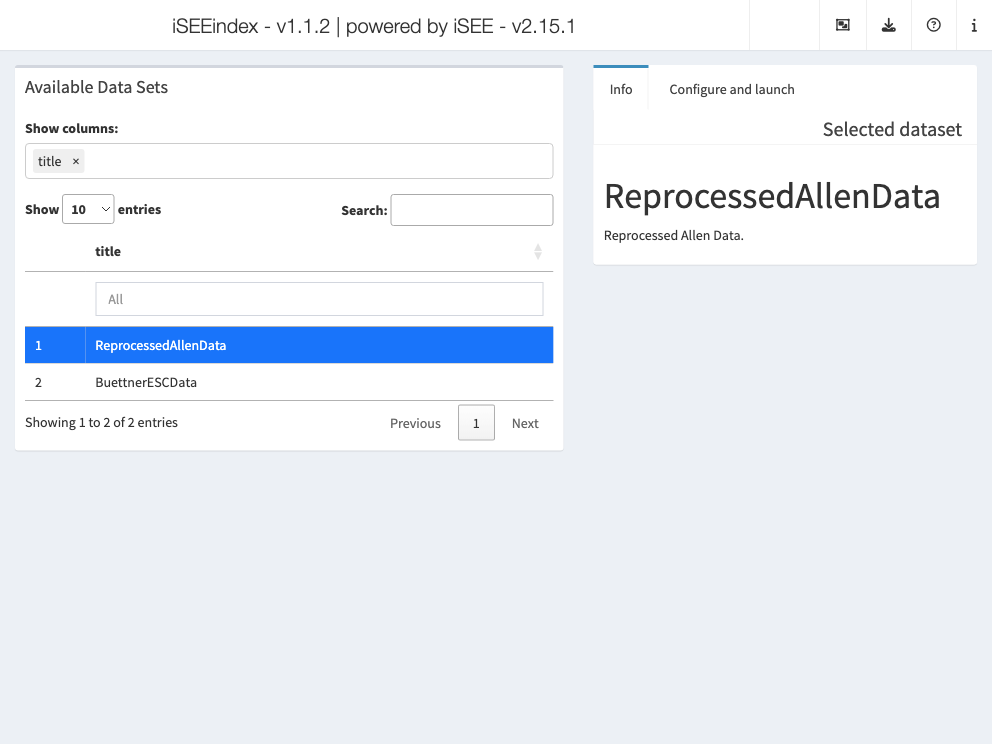
A more elaborate example (referring to the work in (Rigby et al. 2023)) is available at https://rehwinkellab.shinyapps.io/ifnresource/. The source can be found at https://github.com/kevinrue/IFNresource.
Potential use cases can include:
- An app to present and explore the different datasets in your next publication
- An app to explore collection of datasets collaboratively, in consortium-like initiatives
- An app to mirror and enhance the content of e.g. the cellxgene data portal
- Got any ideas on how to use iSEE for such deployments?
Tours: help and storytelling
Tours can be an essential tool to satisfy two needs:
- Helping users to navigate the UI
- Telling a story on an existing configuration
A simple example can be demonstrated with this configuration:
sce_location <- system.file("datasets", "sce_pbmc3k.RDS", package = "iUSEiSEE")
sce_location
#> [1] "/Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/library/iUSEiSEE/datasets/sce_pbmc3k.RDS"
sce <- readRDS(sce_location)
initial_for_tour <- list(
ReducedDimensionPlot(PanelWidth = 3L),
RowDataTable(PanelWidth = 3L),
FeatureAssayPlot(PanelWidth = 3L),
ComplexHeatmapPlot(PanelWidth = 3L)
)This next chunk defines the steps of the tour,
specified by an anchoring point (element) and the content
of that step (intro).
tour <- data.frame(
element = c(
"#Welcome",
"#ReducedDimensionPlot1",
"#RowDataTable1",
"#ComplexHeatmapPlot1",
"#FeatureAssayPlot1",
"#ReducedDimensionPlot1",
"#Conclusion"),
intro = c(
"Welcome to this tour!",
"This is the a reduced dimension plot",
"and this is a table",
"Why not a heatmap?",
"... and now we look at one individual feature.",
"Back to the a reduced dimension plot...",
"Thank you for taking this tour!"),
stringsAsFactors = FALSE
)
app <- iSEE(sce, initial = initial_for_tour, tour = tour)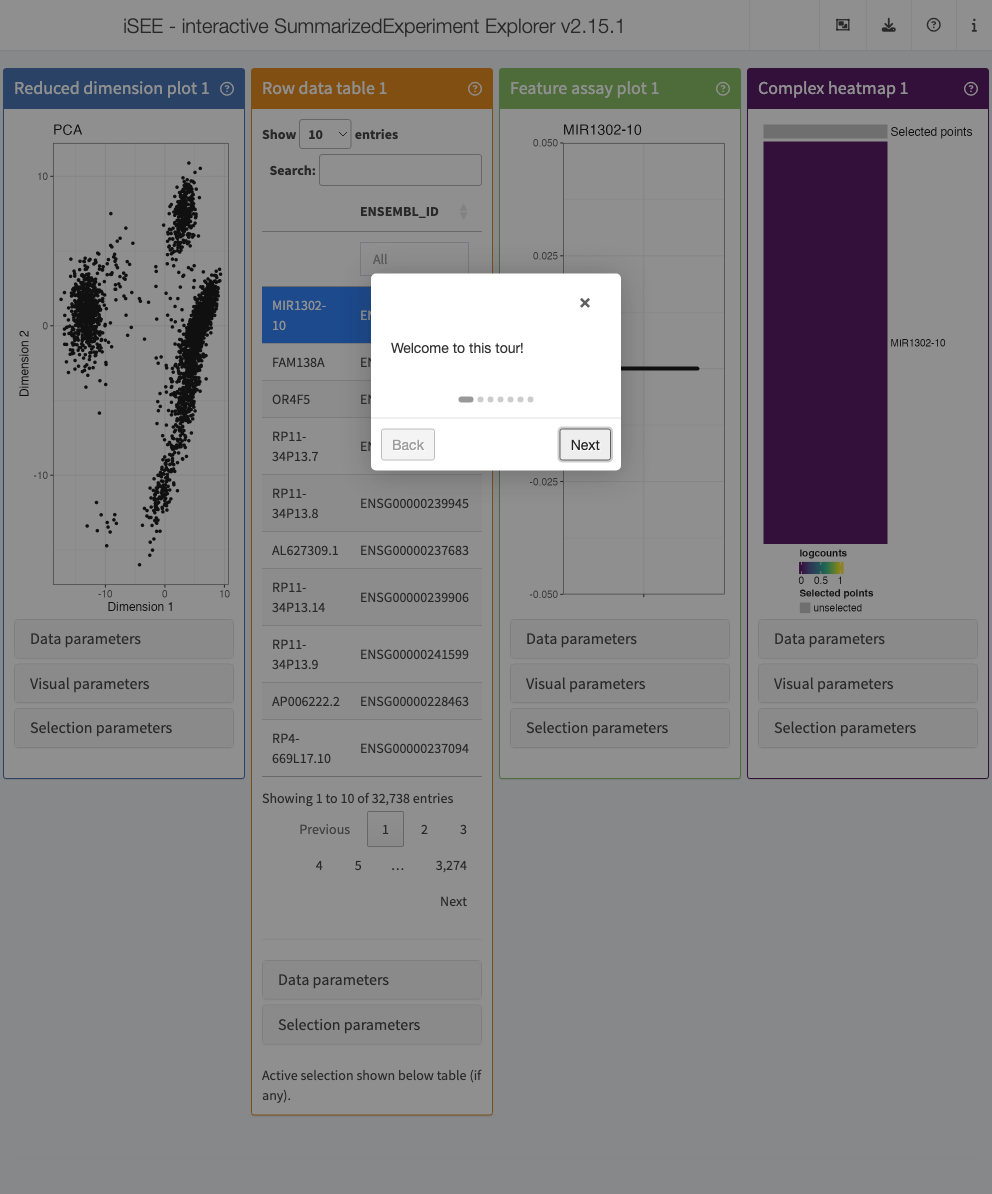
Session info
sessionInfo()
#> R version 4.4.0 (2024-04-24)
#> Platform: x86_64-apple-darwin20
#> Running under: macOS Monterey 12.7.1
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> time zone: Europe/Berlin
#> tzcode source: internal
#>
#> attached base packages:
#> [1] stats4 stats graphics grDevices utils datasets methods
#> [8] base
#>
#> other attached packages:
#> [1] iSEEindex_1.2.0 iSEEhub_1.6.0
#> [3] ExperimentHub_2.12.0 AnnotationHub_3.12.0
#> [5] BiocFileCache_2.12.0 dbplyr_2.5.0
#> [7] iSEEfier_1.0.0 iSEEpathways_1.2.0
#> [9] iSEEde_1.2.0 iSEE_2.15.1
#> [11] SingleCellExperiment_1.26.0 SummarizedExperiment_1.33.3
#> [13] Biobase_2.64.0 GenomicRanges_1.55.4
#> [15] GenomeInfoDb_1.40.0 IRanges_2.38.0
#> [17] S4Vectors_0.42.0 BiocGenerics_0.50.0
#> [19] MatrixGenerics_1.16.0 matrixStats_1.3.0
#> [21] BiocStyle_2.32.0
#>
#> loaded via a namespace (and not attached):
#> [1] RColorBrewer_1.1-3 rstudioapi_0.16.0 jsonlite_1.8.8
#> [4] shape_1.4.6.1 magrittr_2.0.3 farver_2.1.1
#> [7] rmarkdown_2.26 GlobalOptions_0.1.2 fs_1.6.4
#> [10] zlibbioc_1.50.0 ragg_1.3.0 vctrs_0.6.5
#> [13] memoise_2.0.1 RCurl_1.98-1.14 htmltools_0.5.8.1
#> [16] S4Arrays_1.4.0 BiocBaseUtils_1.6.0 curl_5.2.1
#> [19] SparseArray_1.4.0 sass_0.4.9 bslib_0.7.0
#> [22] htmlwidgets_1.6.4 desc_1.4.3 fontawesome_0.5.2
#> [25] listviewer_4.0.0 cachem_1.0.8 igraph_2.0.3
#> [28] mime_0.12 lifecycle_1.0.4 iterators_1.0.14
#> [31] pkgconfig_2.0.3 colourpicker_1.3.0 Matrix_1.7-0
#> [34] R6_2.5.1 fastmap_1.1.1 GenomeInfoDbData_1.2.12
#> [37] shiny_1.8.1.1 clue_0.3-65 digest_0.6.35
#> [40] colorspace_2.1-0 paws.storage_0.5.0 AnnotationDbi_1.65.2
#> [43] DESeq2_1.44.0 RSQLite_2.3.6 textshaping_0.3.7
#> [46] filelock_1.0.3 urltools_1.7.3 fansi_1.0.6
#> [49] httr_1.4.7 abind_1.4-5 mgcv_1.9-1
#> [52] compiler_4.4.0 bit64_4.0.5 withr_3.0.0
#> [55] doParallel_1.0.17 BiocParallel_1.38.0 DBI_1.2.2
#> [58] shinyAce_0.4.2 hexbin_1.28.3 highr_0.10
#> [61] rappdirs_0.3.3 DelayedArray_0.30.0 rjson_0.2.21
#> [64] tools_4.4.0 vipor_0.4.7 httpuv_1.6.15
#> [67] glue_1.7.0 nlme_3.1-164 promises_1.3.0
#> [70] grid_4.4.0 cluster_2.1.6 generics_0.1.3
#> [73] gtable_0.3.5 utf8_1.2.4 XVector_0.44.0
#> [76] stringr_1.5.1 BiocVersion_3.19.1 ggrepel_0.9.5
#> [79] foreach_1.5.2 pillar_1.9.0 limma_3.60.0
#> [82] later_1.3.2 rintrojs_0.3.4 circlize_0.4.16
#> [85] splines_4.4.0 dplyr_1.1.4 lattice_0.22-6
#> [88] bit_4.0.5 paws.common_0.7.2 tidyselect_1.2.1
#> [91] ComplexHeatmap_2.20.0 locfit_1.5-9.9 Biostrings_2.72.0
#> [94] miniUI_0.1.1.1 knitr_1.46 edgeR_4.2.0
#> [97] xfun_0.43 shinydashboard_0.7.2 statmod_1.5.0
#> [100] iSEEhex_1.5.0 DT_0.33 stringi_1.8.3
#> [103] visNetwork_2.1.2 UCSC.utils_1.0.0 yaml_2.3.8
#> [106] shinyWidgets_0.8.6 evaluate_0.23 codetools_0.2-20
#> [109] tibble_3.2.1 BiocManager_1.30.22 cli_3.6.2
#> [112] xtable_1.8-4 systemfonts_1.0.6 munsell_0.5.1
#> [115] jquerylib_0.1.4 iSEEu_1.15.1 Rcpp_1.0.12
#> [118] triebeard_0.4.1 png_0.1-8 parallel_4.4.0
#> [121] blob_1.2.4 pkgdown_2.0.9 ggplot2_3.5.1
#> [124] bitops_1.0-7 viridisLite_0.4.2 scales_1.3.0
#> [127] purrr_1.0.2 crayon_1.5.2 GetoptLong_1.0.5
#> [130] rlang_1.1.3 KEGGREST_1.44.0 shinyjs_2.1.0