Adding a custom header and footer to the landing page
Kevin Rue-Albrecht
University of Oxfordkevin.rue-albrecht@imm.ox.ac.uk
18 October 2024
Source:vignettes/header.Rmd
header.RmdCustom header and footer
Overview
By default, iSEEindex
creates a minimal landing page presenting users with a fluid row that
contains two shinydashboard::box():
- On the left, users can select a data set.
- On the right, users can select an initial configuration and launch the main app interface.
iSEEindex offers the opportunity for app maintainers to add content above (i.e., header) and below (i.e., footer) that row.
That header and footer space may contain any valid UI element that will be displayed in the application UI as is.
Implementation
The function iSEEindex() accepts header and footer
content through its arguments body.header= and
body.footer=.
We recommend wrapping header and footer content in
shiny::fluidRow(), to fit naturally with the main fluid row
(see above).
Example
First, let us set up the core elements of a simple application (see
?iSEEindex).
library("BiocFileCache")
##
# BiocFileCache ----
##
library(BiocFileCache)
bfc <- BiocFileCache(cache = tempdir())
##
# iSEEindex ----
##
dataset_fun <- function() {
x <- yaml::read_yaml(system.file(package = "iSEEindex", "example.yaml"))
x$datasets
}
initial_fun <- function() {
x <- yaml::read_yaml(system.file(package = "iSEEindex", "example.yaml"))
x$initial
}Then, we design header and footer content, each wrapped in a
shiny::fluidRow().
library("shiny")
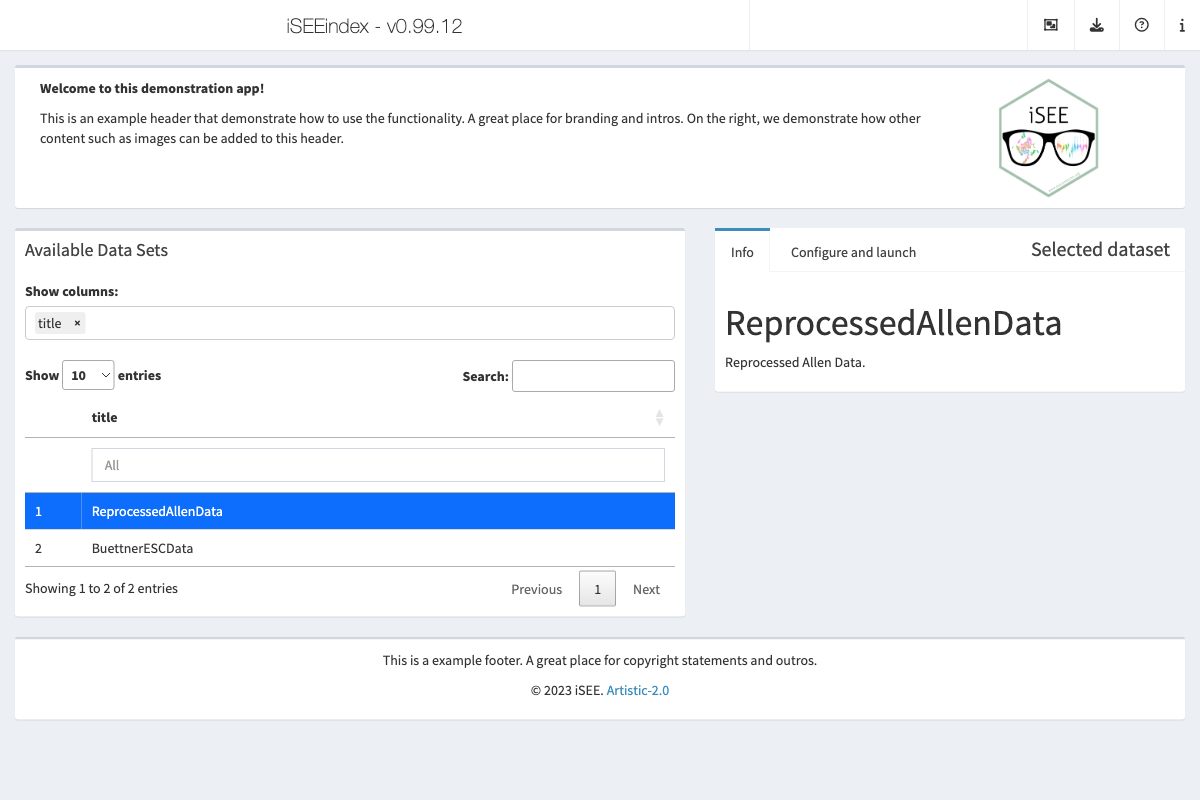
header <- fluidRow(shinydashboard::box(width = 12L,
column(width = 10,
p(strong("Welcome to this demonstration app!")),
p(
"This is an example header that demonstrate how to use the functionality.",
"A great place for branding and intros.",
),
p("On the right, we demonstrate how other content such as images can be added to this header.")
),
column(width = 2, img(src="www/iSEE.png", width = "100px", height = "120px"))
))
footer <- fluidRow(shinydashboard::box(width = 12L,
p("This is a example footer. A great place for copyright statements and outros.", style="text-align: center;"),
p(
"© 2023 iSEE.",
a("Artistic-2.0", href = "https://opensource.org/license/artistic-2-0/"),
style="text-align: center;")
))For the image file to be found, the containing directory must be added to the Shiny resource path as follows:
library("shiny")
addResourcePath("www", system.file(package = "iSEEindex"))Finally, we supply the custom header and footer to
iSEEindex().
library("iSEEindex")
app <- iSEEindex(bfc, dataset_fun, initial_fun,
body.header = header, body.footer = footer
)
if (interactive()) {
shiny::runApp(app, port = 1234)
}
Reproducibility
The iSEEindex package (Rue-Albrecht and Marini, 2024) was made possible thanks to:
- R (R Core Team, 2024)
- BiocStyle (Oleś, 2024)
- knitr (Xie, 2024)
- RefManageR (McLean, 2017)
- rmarkdown (Allaire, Xie, Dervieux, McPherson, Luraschi, Ushey, Atkins, Wickham, Cheng, Chang, and Iannone, 2024)
- sessioninfo (Wickham, Chang, Flight, Müller, and Hester, 2021)
- testthat (Wickham, 2011)
This package was developed using biocthis.
Code for creating the vignette
## Create the vignette
library("rmarkdown")
system.time(render("header.Rmd", "BiocStyle::html_document"))
## Extract the R code
library("knitr")
knit("header.Rmd", tangle = TRUE)Date the vignette was generated.
#> [1] "2024-10-18 10:44:35 UTC"Wallclock time spent generating the vignette.
#> Time difference of 7.396 secsR session information.
#> ─ Session info ───────────────────────────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.4.1 (2024-06-14)
#> os Ubuntu 22.04.5 LTS
#> system x86_64, linux-gnu
#> ui X11
#> language en
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz UTC
#> date 2024-10-18
#> pandoc 3.4 @ /usr/bin/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> abind 1.4-8 2024-09-12 [1] RSPM (R 4.4.0)
#> backports 1.5.0 2024-05-23 [1] RSPM (R 4.4.0)
#> bibtex 0.5.1 2023-01-26 [1] RSPM (R 4.4.0)
#> Biobase * 2.65.1 2024-08-28 [1] Bioconductor 3.20 (R 4.4.1)
#> BiocFileCache * 2.13.2 2024-10-11 [1] Bioconductor 3.20 (R 4.4.1)
#> BiocGenerics * 0.51.3 2024-10-02 [1] Bioconductor 3.20 (R 4.4.1)
#> BiocManager 1.30.25 2024-08-28 [2] CRAN (R 4.4.1)
#> BiocStyle * 2.33.1 2024-06-12 [1] Bioconductor 3.20 (R 4.4.0)
#> bit 4.5.0 2024-09-20 [1] RSPM (R 4.4.0)
#> bit64 4.5.2 2024-09-22 [1] RSPM (R 4.4.0)
#> blob 1.2.4 2023-03-17 [1] RSPM (R 4.4.0)
#> bookdown 0.41 2024-10-16 [1] RSPM (R 4.4.0)
#> bslib 0.8.0 2024-07-29 [2] RSPM (R 4.4.0)
#> cachem 1.1.0 2024-05-16 [2] RSPM (R 4.4.0)
#> circlize 0.4.16 2024-02-20 [1] RSPM (R 4.4.0)
#> cli 3.6.3 2024-06-21 [2] RSPM (R 4.4.0)
#> clue 0.3-65 2023-09-23 [1] RSPM (R 4.4.0)
#> cluster 2.1.6 2023-12-01 [3] CRAN (R 4.4.1)
#> codetools 0.2-20 2024-03-31 [3] CRAN (R 4.4.1)
#> colorspace 2.1-1 2024-07-26 [1] RSPM (R 4.4.0)
#> colourpicker 1.3.0 2023-08-21 [1] RSPM (R 4.4.0)
#> ComplexHeatmap 2.21.1 2024-09-24 [1] Bioconductor 3.20 (R 4.4.1)
#> crayon 1.5.3 2024-06-20 [2] RSPM (R 4.4.0)
#> curl 5.2.3 2024-09-20 [2] RSPM (R 4.4.0)
#> DBI 1.2.3 2024-06-02 [1] RSPM (R 4.4.0)
#> dbplyr * 2.5.0 2024-03-19 [1] RSPM (R 4.4.0)
#> DelayedArray 0.31.14 2024-10-03 [1] Bioconductor 3.20 (R 4.4.1)
#> desc 1.4.3 2023-12-10 [2] RSPM (R 4.4.0)
#> digest 0.6.37 2024-08-19 [2] RSPM (R 4.4.0)
#> doParallel 1.0.17 2022-02-07 [1] RSPM (R 4.4.0)
#> dplyr 1.1.4 2023-11-17 [1] RSPM (R 4.4.0)
#> DT 0.33 2024-04-04 [1] RSPM (R 4.4.0)
#> evaluate 1.0.1 2024-10-10 [2] RSPM (R 4.4.0)
#> fansi 1.0.6 2023-12-08 [2] RSPM (R 4.4.0)
#> fastmap 1.2.0 2024-05-15 [2] RSPM (R 4.4.0)
#> filelock 1.0.3 2023-12-11 [1] RSPM (R 4.4.0)
#> fontawesome 0.5.2 2023-08-19 [2] RSPM (R 4.4.0)
#> foreach 1.5.2 2022-02-02 [1] RSPM (R 4.4.0)
#> fs 1.6.4 2024-04-25 [2] RSPM (R 4.4.0)
#> generics 0.1.3 2022-07-05 [1] RSPM (R 4.4.0)
#> GenomeInfoDb * 1.41.2 2024-10-02 [1] Bioconductor 3.20 (R 4.4.1)
#> GenomeInfoDbData 1.2.13 2024-10-18 [1] Bioconductor
#> GenomicRanges * 1.57.2 2024-10-09 [1] Bioconductor 3.20 (R 4.4.1)
#> GetoptLong 1.0.5 2020-12-15 [1] RSPM (R 4.4.0)
#> ggplot2 3.5.1 2024-04-23 [1] RSPM (R 4.4.0)
#> ggrepel 0.9.6 2024-09-07 [1] RSPM (R 4.4.0)
#> GlobalOptions 0.1.2 2020-06-10 [1] RSPM (R 4.4.0)
#> glue 1.8.0 2024-09-30 [2] RSPM (R 4.4.0)
#> gtable 0.3.5 2024-04-22 [1] RSPM (R 4.4.0)
#> highr 0.11 2024-05-26 [2] RSPM (R 4.4.0)
#> htmltools 0.5.8.1 2024-04-04 [2] RSPM (R 4.4.0)
#> htmlwidgets 1.6.4 2023-12-06 [2] RSPM (R 4.4.0)
#> httpuv 1.6.15 2024-03-26 [2] RSPM (R 4.4.0)
#> httr 1.4.7 2023-08-15 [1] RSPM (R 4.4.0)
#> igraph 2.0.3 2024-03-13 [1] RSPM (R 4.4.0)
#> IRanges * 2.39.2 2024-07-17 [1] Bioconductor 3.20 (R 4.4.1)
#> iSEE 2.17.4 2024-09-03 [1] Bioconductor 3.20 (R 4.4.1)
#> iSEEindex * 1.3.3 2024-10-18 [1] Bioconductor
#> iterators 1.0.14 2022-02-05 [1] RSPM (R 4.4.0)
#> jquerylib 0.1.4 2021-04-26 [2] RSPM (R 4.4.0)
#> jsonlite 1.8.9 2024-09-20 [2] RSPM (R 4.4.0)
#> knitr 1.48 2024-07-07 [2] RSPM (R 4.4.0)
#> later 1.3.2 2023-12-06 [2] RSPM (R 4.4.0)
#> lattice 0.22-6 2024-03-20 [3] CRAN (R 4.4.1)
#> lifecycle 1.0.4 2023-11-07 [2] RSPM (R 4.4.0)
#> listviewer 4.0.0 2023-09-30 [1] RSPM (R 4.4.0)
#> lubridate 1.9.3 2023-09-27 [1] RSPM (R 4.4.0)
#> magrittr 2.0.3 2022-03-30 [2] RSPM (R 4.4.0)
#> Matrix 1.7-0 2024-04-26 [3] CRAN (R 4.4.1)
#> MatrixGenerics * 1.17.0 2024-05-01 [1] Bioconductor 3.20 (R 4.4.0)
#> matrixStats * 1.4.1 2024-09-08 [1] RSPM (R 4.4.0)
#> memoise 2.0.1 2021-11-26 [2] RSPM (R 4.4.0)
#> mgcv 1.9-1 2023-12-21 [3] CRAN (R 4.4.1)
#> mime 0.12 2021-09-28 [2] RSPM (R 4.4.0)
#> miniUI 0.1.1.1 2018-05-18 [2] RSPM (R 4.4.0)
#> munsell 0.5.1 2024-04-01 [1] RSPM (R 4.4.0)
#> nlme 3.1-166 2024-08-14 [2] RSPM (R 4.4.0)
#> paws.common 0.7.7 2024-10-03 [1] RSPM (R 4.4.0)
#> paws.storage 0.7.0 2024-09-11 [1] RSPM (R 4.4.0)
#> pillar 1.9.0 2023-03-22 [2] RSPM (R 4.4.0)
#> pkgconfig 2.0.3 2019-09-22 [2] RSPM (R 4.4.0)
#> pkgdown 2.1.1 2024-09-17 [2] RSPM (R 4.4.0)
#> plyr 1.8.9 2023-10-02 [1] RSPM (R 4.4.0)
#> png 0.1-8 2022-11-29 [1] RSPM (R 4.4.0)
#> promises 1.3.0 2024-04-05 [2] RSPM (R 4.4.0)
#> purrr 1.0.2 2023-08-10 [2] RSPM (R 4.4.0)
#> R6 2.5.1 2021-08-19 [2] RSPM (R 4.4.0)
#> ragg 1.3.3 2024-09-11 [2] RSPM (R 4.4.0)
#> RColorBrewer 1.1-3 2022-04-03 [1] RSPM (R 4.4.0)
#> Rcpp 1.0.13 2024-07-17 [2] RSPM (R 4.4.0)
#> RefManageR * 1.4.0 2022-09-30 [1] RSPM (R 4.4.0)
#> rintrojs 0.3.4 2024-01-11 [1] RSPM (R 4.4.0)
#> rjson 0.2.23 2024-09-16 [1] RSPM (R 4.4.0)
#> rlang 1.1.4 2024-06-04 [2] RSPM (R 4.4.0)
#> rmarkdown 2.28 2024-08-17 [2] RSPM (R 4.4.0)
#> RSQLite 2.3.7 2024-05-27 [1] RSPM (R 4.4.0)
#> S4Arrays 1.5.11 2024-10-14 [1] Bioconductor 3.20 (R 4.4.1)
#> S4Vectors * 0.43.2 2024-07-17 [1] Bioconductor 3.20 (R 4.4.1)
#> sass 0.4.9 2024-03-15 [2] RSPM (R 4.4.0)
#> scales 1.3.0 2023-11-28 [1] RSPM (R 4.4.0)
#> sessioninfo * 1.2.2 2021-12-06 [2] RSPM (R 4.4.0)
#> shape 1.4.6.1 2024-02-23 [1] RSPM (R 4.4.0)
#> shiny * 1.9.1 2024-08-01 [2] RSPM (R 4.4.0)
#> shinyAce 0.4.2 2022-05-06 [1] RSPM (R 4.4.0)
#> shinydashboard 0.7.2 2021-09-30 [1] RSPM (R 4.4.0)
#> shinyjs 2.1.0 2021-12-23 [1] RSPM (R 4.4.0)
#> shinyWidgets 0.8.7 2024-09-23 [1] RSPM (R 4.4.0)
#> SingleCellExperiment * 1.27.2 2024-05-24 [1] Bioconductor 3.20 (R 4.4.0)
#> SparseArray 1.5.44 2024-10-06 [1] Bioconductor 3.20 (R 4.4.1)
#> stringi 1.8.4 2024-05-06 [2] RSPM (R 4.4.0)
#> stringr 1.5.1 2023-11-14 [2] RSPM (R 4.4.0)
#> SummarizedExperiment * 1.35.4 2024-10-09 [1] Bioconductor 3.20 (R 4.4.1)
#> systemfonts 1.1.0 2024-05-15 [2] RSPM (R 4.4.0)
#> textshaping 0.4.0 2024-05-24 [2] RSPM (R 4.4.0)
#> tibble 3.2.1 2023-03-20 [2] RSPM (R 4.4.0)
#> tidyselect 1.2.1 2024-03-11 [1] RSPM (R 4.4.0)
#> timechange 0.3.0 2024-01-18 [1] RSPM (R 4.4.0)
#> triebeard 0.4.1 2023-03-04 [1] RSPM (R 4.4.0)
#> UCSC.utils 1.1.0 2024-05-01 [1] Bioconductor 3.20 (R 4.4.0)
#> urltools 1.7.3 2019-04-14 [1] RSPM (R 4.4.0)
#> utf8 1.2.4 2023-10-22 [2] RSPM (R 4.4.0)
#> vctrs 0.6.5 2023-12-01 [2] RSPM (R 4.4.0)
#> vipor 0.4.7 2023-12-18 [1] RSPM (R 4.4.0)
#> viridisLite 0.4.2 2023-05-02 [1] RSPM (R 4.4.0)
#> xfun 0.48 2024-10-03 [2] RSPM (R 4.4.0)
#> xml2 1.3.6 2023-12-04 [2] RSPM (R 4.4.0)
#> xtable 1.8-4 2019-04-21 [2] RSPM (R 4.4.0)
#> XVector 0.45.0 2024-05-01 [1] Bioconductor 3.20 (R 4.4.0)
#> yaml 2.3.10 2024-07-26 [2] RSPM (R 4.4.0)
#> zlibbioc 1.51.1 2024-06-05 [1] Bioconductor 3.20 (R 4.4.0)
#>
#> [1] /__w/_temp/Library
#> [2] /usr/local/lib/R/site-library
#> [3] /usr/local/lib/R/library
#>
#> ──────────────────────────────────────────────────────────────────────────────────────────────────Bibliography
This vignette was generated using BiocStyle (Oleś, 2024) with knitr (Xie, 2024) and rmarkdown (Allaire, Xie, Dervieux et al., 2024) running behind the scenes.
Citations made with RefManageR (McLean, 2017).
[1] J. Allaire, Y. Xie, C. Dervieux, et al. rmarkdown: Dynamic Documents for R. R package version 2.28. 2024. URL: https://github.com/rstudio/rmarkdown.
[2] M. W. McLean. “RefManageR: Import and Manage BibTeX and BibLaTeX References in R”. In: The Journal of Open Source Software (2017). DOI: 10.21105/joss.00338.
[3] A. Oleś. BiocStyle: Standard styles for vignettes and other Bioconductor documents. R package version 2.33.1. 2024. DOI: 10.18129/B9.bioc.BiocStyle. URL: https://bioconductor.org/packages/BiocStyle.
[4] R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria, 2024. URL: https://www.R-project.org/.
[5] K. Rue-Albrecht and F. Marini. iSEEindex: iSEE extension for a landing page to a custom collection of data sets. R package version 1.3.3. 2024. URL: https://github.com/iSEE/iSEEindex.
[6] H. Wickham. “testthat: Get Started with Testing”. In: The R Journal 3 (2011), pp. 5–10. URL: https://journal.r-project.org/archive/2011-1/RJournal_2011-1_Wickham.pdf.
[7] H. Wickham, W. Chang, R. Flight, et al. sessioninfo: R Session Information. R package version 1.2.2, https://r-lib.github.io/sessioninfo/. 2021. URL: https://github.com/r-lib/sessioninfo#readme.
[8] Y. Xie. knitr: A General-Purpose Package for Dynamic Report Generation in R. R package version 1.48. 2024. URL: https://yihui.org/knitr/.